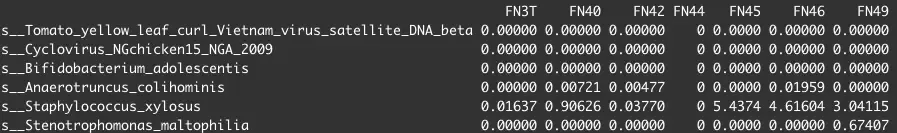
WX20201110-151431.png
以及样本的各种信息sample.info,形如:
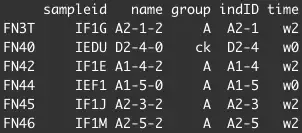
WX20201110-151645.png
- 步骤1:多滤掉低丰度数据
#过滤掉物种丰度为0的样本数过多的行
> filter.index <- apply(taxa.raw, 1, function(X){sum(X>0)>0.1*length(X)})
> taxa.filter <- taxa.raw[filter.index, ]
#确保样本丰度和相加为100
> taxa.filter <- 100*sweep(taxa.filter, 2, colSums(taxa.filter), FUN="/")
> taxa.data <- as.data.frame(t(taxa.filter))
- 步骤2:创建协变量矩阵
我们可以用如下代码为group、time以及group和time的交互项创建协变量矩阵:
> reg.cov <- tibble::rownames_to_column(sample.info, var = 'Sample') %>%
dplyr::select(Sample, group, indID, time) %>%
dplyr::mutate(time = ifelse(time == 'w0', 0, ifelse(time == 'w2', 1, ifelse(time == 'w4', 2,NA)))) %>% #创建时间变量代码:w0是0,w2是1,w4是2;
dplyr::mutate(group = ifelse(group == 'ck', 0, 1)) %>% #创建分组变量代码:ck组为0,A组为1
dplyr::mutate(timeXgroup = time*group) %>% #创建时间与分组交互代码
dplyr::mutate(indID = as.character(indID)) %>%
as.data.frame
#过滤掉采样不全的个体数据
> ind.count <- table(reg.cov$indID)
> reg.cov <- subset(reg.cov, indID %in% names(ind.count)[ind.count == 3])
> head(reg.cov)
Sample group indID time timeXgroup
1 FN3T 1 A2-1 1 1
2 FN40 0 D2-4 0 0
3 FN42 1 A1-4 1 1
4 FN44 1 A1-5 0 0
5 FN45 1 A2-3 1 1
7 FN49 0 D2-2 1 0
- 步骤3:拟合模型
先将w0和w1+w2的数据分别取出备用:
> reg.cov.w0 <- subset(reg.cov, time == 0)
> rownames(reg.cov.w0) <- reg.cov.w0$indID
> head(reg.cov.w0)
Sample group indID time timeXgroup
D2-4 FN40 0 D2-4 0 0
A1-5 FN44 1 A1-5 0 0
D2-5 FN4D 0 D2-5 0 0
D1-2 FN54 0 D1-2 0 0
A1-4 FN5J 1 A1-4 0 0
A2-4 FN60 1 A2-4 0 0
> reg.cov.w12 <- subset(reg.cov, time != 0)
> reg.cov.w12 <- na.omit(data.frame(baseline.sample = reg.cov.w0[reg.cov.w12$indID, 'Sample'],
baseline.subject = reg.cov.w0[reg.cov.w12$indID, 'indID'],
reg.cov.w12,
stringsAsFactors = F))
> head(reg.cov.w12)
baseline.sample baseline.subject Sample group indID time timeXgroup
1 FN70 A2-1 FN3T 1 A2-1 1 1
3 FN5J A1-4 FN42 1 A1-4 1 1
5 FN68 A2-3 FN45 1 A2-3 1 1
7 FN71 D2-2 FN49 0 D2-2 1 0
8 FN54 D1-2 FN4A 0 D1-2 2 0
9 FN6T D1-5 FN4B 0 D1-5 1 0
然后单独为每一个物种进行拟合(为了便于说明,此处我们仅以其中一个物种丰度举例:
library(dplyr)
library(nlme)
> taxa.all <- colnames(taxa.data)
> p.taxa.list.lme <- list()
> spe <- taxa.all[9]
> spe
[1] "s__Bacteroides_vulgatus"
> X <- data.frame(Baseline = taxa.data[reg.cov.w12$baseline.sample, spe]/100,
reg.cov.w12[, c('time', 'group')])
> rownames(X) <- reg.cov.w12$Sample
> head(X)
Baseline time group
FN3T 0.0004122127 1 1
FN42 0.0001939168 1 1
FN45 0.0026208886 1 1
FN49 0.0003091166 1 0
FN4A 0.0007188949 2 0
FN4B 0.0005986076 1 0
> Z <- X
> subject.ind <- reg.cov.w12$indID
> time.ind <- reg.cov.w12$time
> Y <- taxa.data[reg.cov.w12$Sample, spe]/100
> cat('Zeros/All',sum(Y==0),'/',length(Y),'\n')
Zeros/All 0 / 30
#对Y进行变换(先开方在反正弦?)
> tdata <- data.frame(Y.tran = asin(sqrt(Y)), X, SID = subject.ind)
> head(tdata)
Y.tran Baseline time group SID
FN3T 0.04531814 0.0004122127 1 1 A2-1
FN42 0.05269246 0.0001939168 1 1 A1-4
FN45 0.01527299 0.0026208886 1 1 A2-3
FN49 0.02599619 0.0003091166 1 0 D2-2
FN4A 0.01817293 0.0007188949 2 0 D1-2
FN4B 0.03147346 0.0005986076 1 0 D1-5
#进行拟合(以Baseline、time以及group为固定效应,以SID为随机效应)
> lme.fit <- try(lme(Y.tran ~ Baseline + time + group, random = ~1|SID, data = tdata))
随机效应的表达方式:
(1)1|SID:SID是随机截距,Baseline、time、group是固定斜率,也就是他们前面的参数是固定的;
(2)0 + time|SID:SID是固定截距,time是随机斜率,Baseline和time是固定斜率;
(3)1 + time|SID:SID是随机截距,time是随机斜率,Baseline和time是固定斜率;
> summary(lme.fit)
...
Random effects:
Formula: ~1 | SID
(Intercept) Residual
StdDev: 0.007468231 0.009009141
Fixed effects: Y.tran ~ Baseline + time + group
Value Std.Error DF t-value p-value
(Intercept) 0.0236327 0.0060813 14 3.886108 0.0016
Baseline -1.2738533 1.3537572 12 -0.940976 0.3653
time 0.0023881 0.0032897 14 0.725932 0.4798
group 0.0016992 0.0053765 12 0.316037 0.7574
...
这样我们就得到了每个自变量的参数,以及P值,后续我们再使用多重检验矫正就可以得到矫正后的P值了。
文章均来自互联网如有不妥请联系作者删除QQ:314111741 地址:http://www.mqs.net/post/14340.html

添加新评论